Home > Mutagenetix
The Mutagenetix website serves to record and disseminate information on the mouse phenotypes and mutations generated in the Center for the Genetics of Host Defense. The Phenotypic Mutations and Incidental Mutations may simply be browsed in list form. Importantly, the site also provides a variety of sorting, filtering, and searching functionalities to enable rapid identification of specific genes, phenotypes, or mutations of interest to the individual researcher. The site is regularly updated, with new mutation and phenotype data added weekly. Key features of the Mutagenetix website are described below.
Phenotypic Mutations
The Phenotypic Mutations page (Figure 1) lists all mutations with statistically significant linkage to one or more phenotypes identified during phenotypic screening. Selecting a particular mutation will open the mutation record, which contains researched and referenced background information and a putative or confirmed mechanism of the mutation, along with detailed illustrations. Phenotypic Mutation records automatically display a protein domain diagram predicted by SMART and the position of the mutation therein, and for missense mutations, the predicted severity of protein damage caused by the mutation as assessed by PolyPhen-2. Each record provides links to the MGI (Mouse Genome Informatics) and Ensemble database records for the gene, to literature sources, and to the repository from which the mouse strain can be obtained, if available. A genotyping protocol or computationally predicted primer sequences for genotyping are provided.
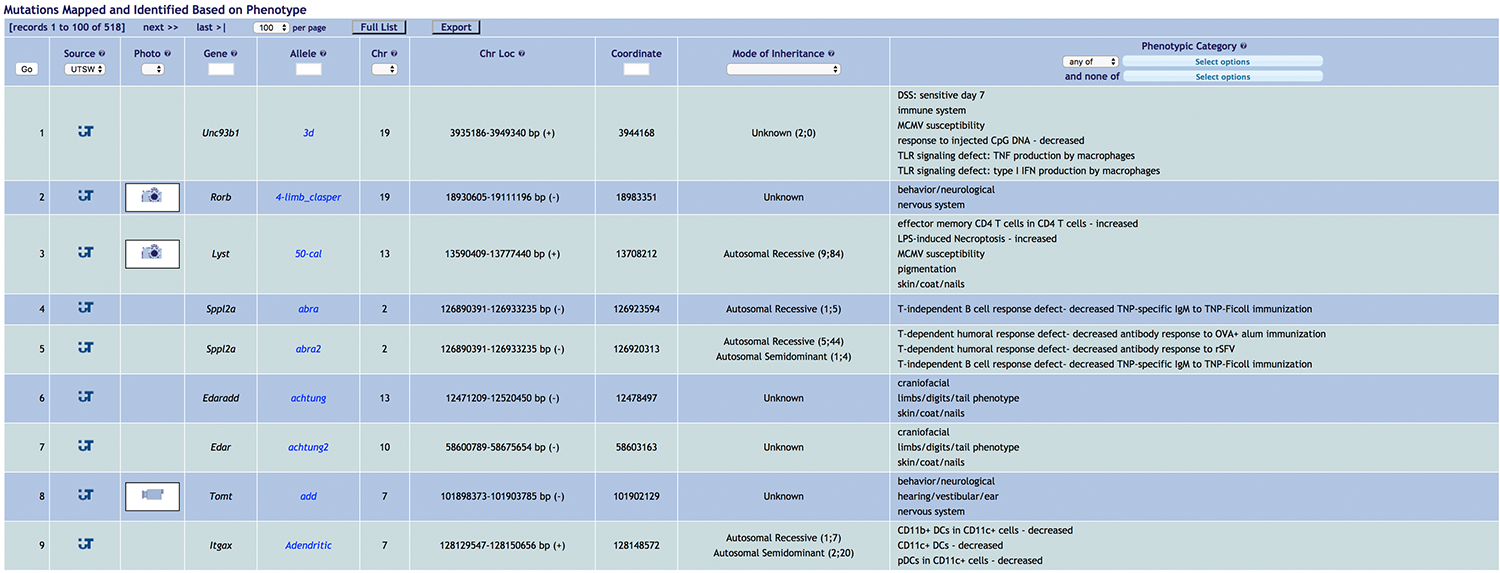 Figure 1. The Phenotypic Mutations list. The list may be sorted by each column heading (Gene, Allele, Chromosome, Chromosome Location, Coordinate, Mode of Inheritance, Phenotypic Category, and others), and filtered by making specifications in several columns (Gene, Allele, Chromosome, Coordinate, Mode of Inheritance, Phenotypic Category).
Figure 1. The Phenotypic Mutations list. The list may be sorted by each column heading (Gene, Allele, Chromosome, Chromosome Location, Coordinate, Mode of Inheritance, Phenotypic Category, and others), and filtered by making specifications in several columns (Gene, Allele, Chromosome, Coordinate, Mode of Inheritance, Phenotypic Category).Selecting a particular mutation will open the mutation record, which is researched and written by the indicated Science Writer(s), who discusses each strain with the PI as well as students and/or postdocs, listed as Authors, who made the initial phenotypic discovery and performed all experimental characterization. Referenced background information and a putative or confirmed mechanism of the mutation are given, along with detailed illustrations. Phenotypic Mutation records automatically display a protein domain diagram predicted by SMART and the position of the mutation therein, and the predicted severity of protein damage caused by the mutation as assessed by PolyPhen-2. Each record provides links to the MGI (Mouse Genome Informatics) and Ensemble database records for the gene, to literature sources, and to the repository from which the mouse strain can be obtained, if available. A genotyping protocol or computationally predicted primer sequences for genotyping are provided.
Incidental Mutations
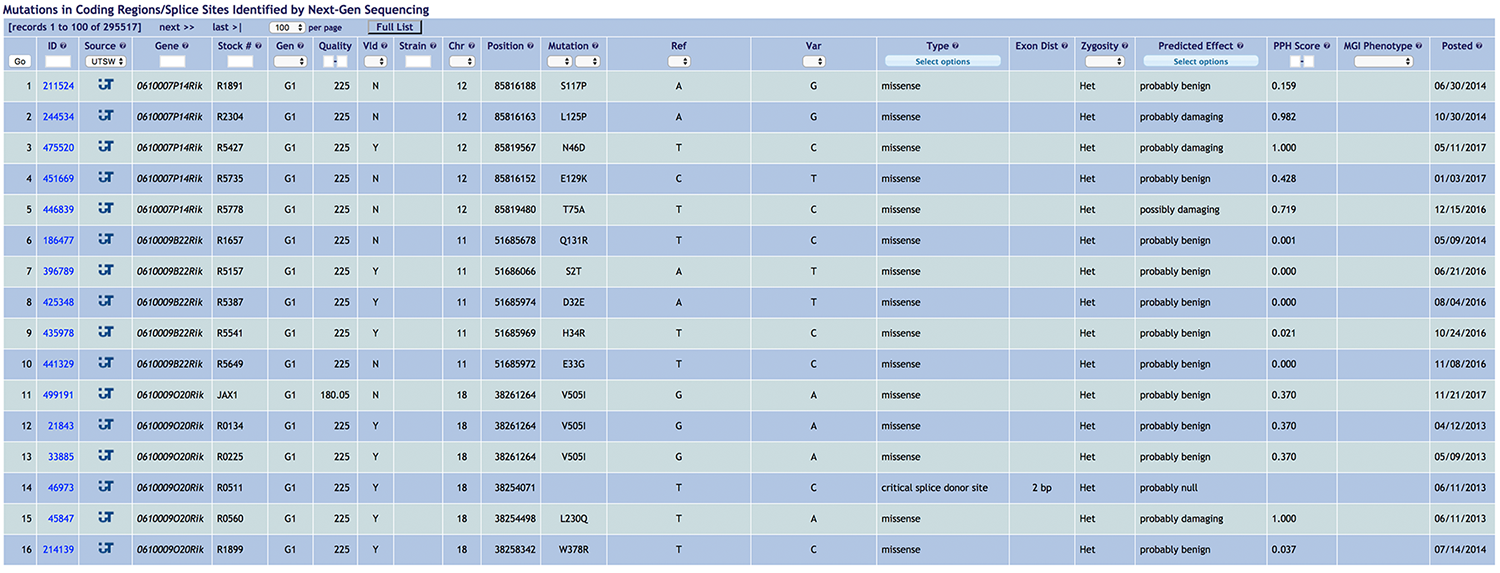
Our Technical Approach to forward genetic analysis involves exome sequencing of the G1 male progenitor of each pedigree carrying ENU-induced mutations. On average, 97% of all coding and critical splice junction nucleotides are sequenced with ≥10X coverage using an Illumina HiSeq 2500 platform. Mutations are validated by genotyping the G1, G2, and G3 mice. The Incidental Mutations page lists all mutations identified and validated in this manner, some of which may also appear in the Phenotypic Mutations list. Selecting a particular mutation will open the mutation record, which is automatically populated with a protein domain diagram predicted by SMART and the position of the mutation therein, the predicted severity of protein damage caused by the mutation as assessed by PolyPhen-2, and links to the MGI (Mouse Genome Informatics) and Ensemble database records for the gene, among other information. Also provided are a list of other mutations found in the strain, a list of other alleles of the mutated gene, and computationally predicted primer sequences to be used for genotyping.
Linkage Explorer (NEW! Coming 2018)
The Linkage Explorer application is used to search and display linkage data. A researcher may search for phenotypes (among those screened in the Center for the Genetics of Host Defense) linked to a gene of interest, or conversely, for mutated genes linked to a phenotype of interest. With a comprehensive collection of search functions, Linkage Explorer searches can be refined to focus on the mutations of greatest relevance. Please read the section Automated Mutation Identification on the Technical Approach page for a detailed description of Linkage Explorer and its capabilities.
Mutation Statistics
The Mutation Statistics page displays automatically updated statistics on the mutation types, DNA base changes, and amino acid changes created by ENU mutagenesis in the Beutler Laboratory since 2001.
Request Mice
All mutant mouse strains produced in the Beutler Laboratory are available to the scientific community. For those not available from a public repository, the Request Mice page provides instructions for requesting them directly from the Beutler Laboratory.